A wrapper for pls2b() specifically aimed at summarising
covariation between shape data and other external, non-shape variable(s).
pls_shapes(X, shapes, tree = NULL, LOOCV = FALSE, recompute = FALSE)Arguments
- X
A matrix with variables as columns and observations as rows, representing the external variables that supervise ordination (corresponding to the first block).
- shapes
Shape data (corresponding to the second block of
pls2b.- tree
A
"phylo"object containing a phylogenetic tree. Tip labels should match the row number and names fromxandy.- LOOCV
Logical; whether to apply leave-one-out cross-validation.
- recompute
Logical; whether to re-compute rotation matrix using the scores resulting from LOOCV.
Value
A "pls_shapes" or "phy_pls_shapes" object formatted
following the "prcomp" class:
$x:the scores from the shapes block.$x2:the scores from the supervising block (i.e., thexscores.$sdev:the standard deviations of the PLS axes (i.e., the standard deviations calculated from the scores of each axis in the shape block).$rotation:a matrix of vector coefficients for the shape block.$center:the mean values of the original variables from the shape block.$totvar:the sum of the variances of the original variables in the shape block.
Details
This function finds the linear combination maximizing covariation
between a block of variables assumed to be shape variables formatted as
a 2-margins matrix and another block which can be either another set of
shape variables or one or more non-shape variables for supervising the
analysis. If a phylogenetic tree is supplied, observations from x
and shapes blocks are treated as data measured for its tips. In this
case, the resulting axes maximize the residual covariation among blocks
left after removing covariation among blocks accounted by phylogenetic
history (assuming a Brownian model of evolution and 100% phylogenetic
signal).
It has been reported that PLS (as an algebraic equivalent of bgPCA) produces spurious covariation between blocks when the number of variables exceeds the number of observations (which is a common situation in geometric morphometrics analyses). This problem can be alleviated by carrying out a leave-one-out cross-validation (LOOCV; i.e., each observation is excluded from the calculation of PLS axes before its projection in the resulting ordination as a way to calculate its score).
The number of axes in both blocks will be equal to the number of variables in the smallest block (i.e., the one with less variables), unless the number of observations is lower (in which case this will be equal to the number PLS axes).
References
Rohlf, F. J., & Corti, M. (2000). Use of two-block partial least-squares to study covariation in shape. Systematic Biology, 49(4), 740-753.
Zelditch, M. L., Swiderski, D. L., & Sheets, H. D. (2012). Geometric morphometrics for biologists: A primer. 2nd ed. Academic Press.
Bookstein, F. L. (2019). Pathologies of between-groups principal components analysis in geometric morphometrics. Evolutionary Biology, 46(4), 271-302.
See also
Examples
#load data
data("tails")
#extract shapes and sizes, compute mean shapes and sizes for all species,
#extract tree
shapes <- tails$shapes
sizes <- log(tails$sizes)
sp_shapes <- expected_shapes(shapes, tails$data$species)
sp_sizes <- tapply(X = log(sizes), INDEX = tails$data$species, FUN = mean)
tree <- tails$tree
#perform PLS between shape and size
pls <- pls_shapes(shapes = shapes, X = sizes)
#inspect results
names(pls) #the contents of the resulting object
#> [1] "sdev" "rotation" "x" "x2" "center" "totvar"
exp_var(pls) #variance explained by each axis
#> variance cummulative
#> PLS-1 74.05123 74.05123
#> PLS-2 NA NA
#> PLS-3 NA NA
#> PLS-4 NA NA
#> PLS-5 NA NA
#> PLS-6 NA NA
#> PLS-7 NA NA
#> PLS-8 NA NA
#> PLS-9 NA NA
#> PLS-10 NA NA
plot(pls$x2, pls$x) #ordination
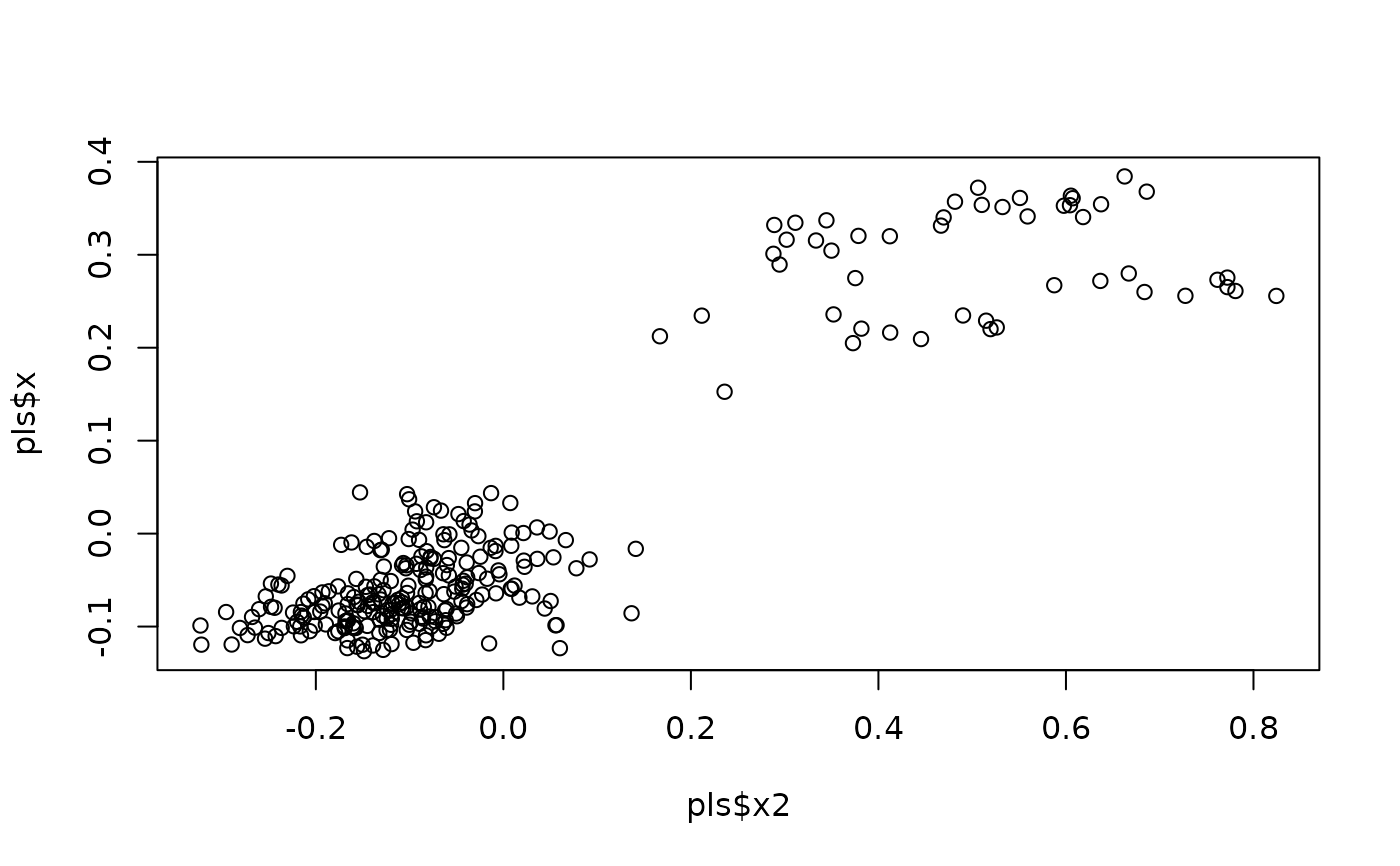 #perform PLS between shape and size with leave-one-out CV
pls_cv <- pls_shapes(shapes = shapes, X = sizes, LOOCV = TRUE)
#inspect results
names(pls_cv) #the contents of the resulting object
#> [1] "sdev" "rotation" "x" "x2" "center" "totvar"
exp_var(pls_cv) #variance explained by each axis
#> variance cummulative
#> PLS-1 73.48807 73.48807
#> PLS-2 NA NA
#> PLS-3 NA NA
#> PLS-4 NA NA
#> PLS-5 NA NA
#> PLS-6 NA NA
#> PLS-7 NA NA
#> PLS-8 NA NA
#> PLS-9 NA NA
#> PLS-10 NA NA
plot(pls_cv$x2, pls_cv$x) #ordination
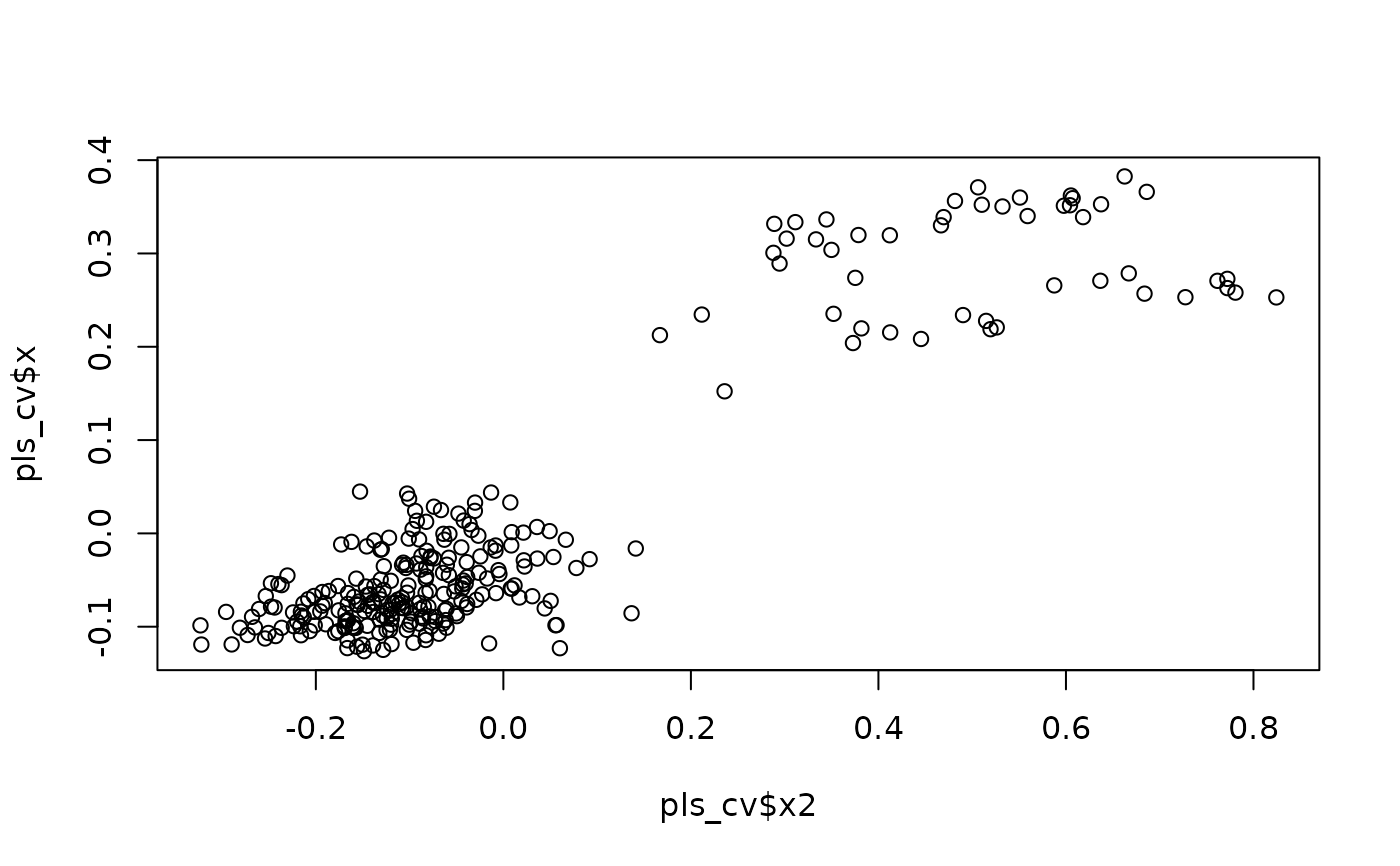
#perform PLS between shape and size with leave-one-out CV
pls_cv <- pls_shapes(shapes = shapes, X = sizes, LOOCV = TRUE)
#inspect results
names(pls_cv) #the contents of the resulting object
#> [1] "sdev" "rotation" "x" "x2" "center" "totvar"
exp_var(pls_cv) #variance explained by each axis
#> variance cummulative
#> PLS-1 73.48807 73.48807
#> PLS-2 NA NA
#> PLS-3 NA NA
#> PLS-4 NA NA
#> PLS-5 NA NA
#> PLS-6 NA NA
#> PLS-7 NA NA
#> PLS-8 NA NA
#> PLS-9 NA NA
#> PLS-10 NA NA
plot(pls_cv$x2, pls_cv$x) #ordination
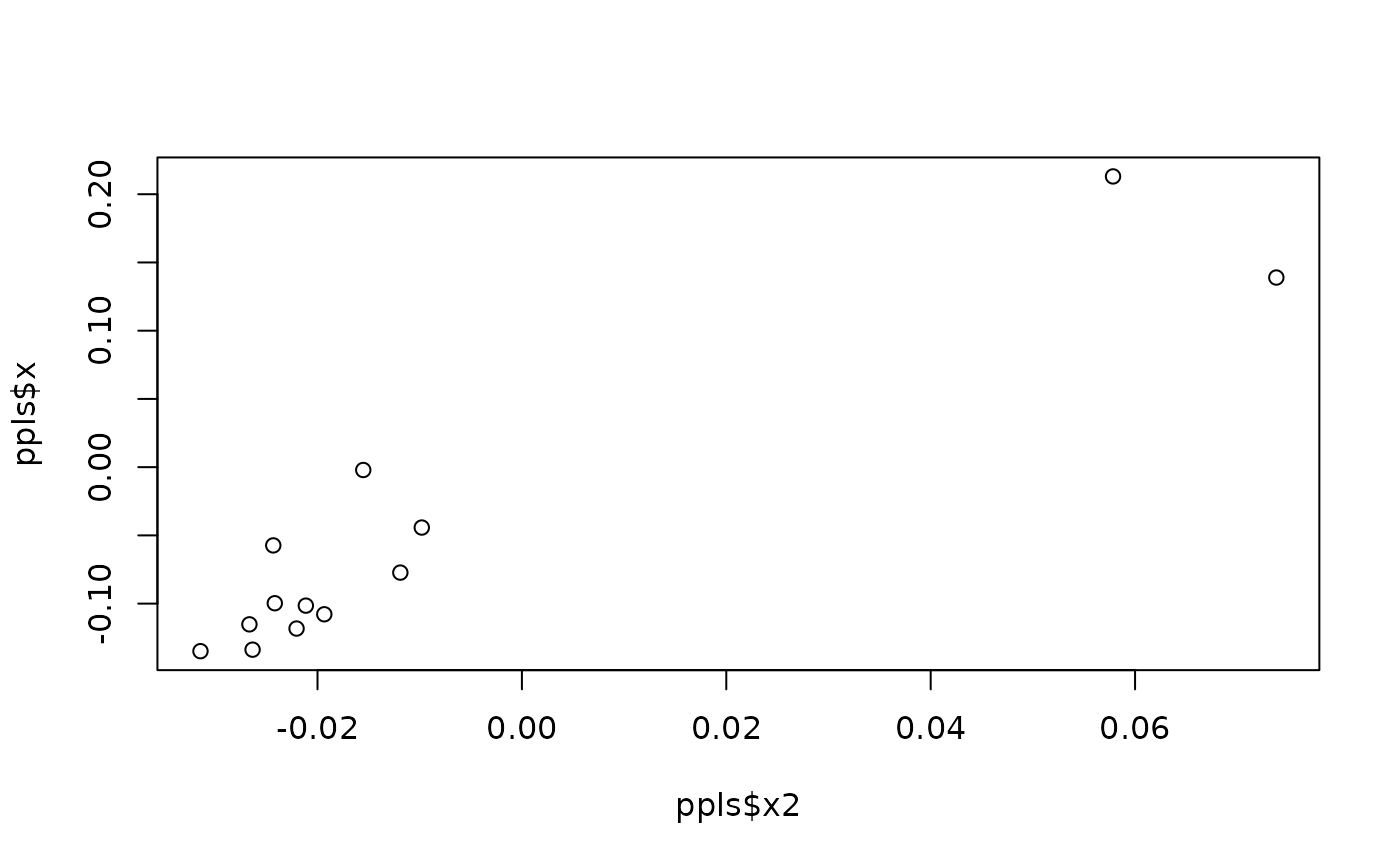
 #perform phylogenetic PLS between shape and size (for species means)
ppls <- pls_shapes(shapes = sp_shapes, X = sp_sizes, tree = tree)
#inspect results
names(ppls) #the contents of the resulting object
#> [1] "sdev" "rotation" "x" "x2" "center" "totvar"
exp_var(ppls) #variance explained by each axis
#> variance cummulative
#> phyPLS-1 75.10454 75.10454
#> phyPLS-2 NA NA
#> phyPLS-3 NA NA
#> phyPLS-4 NA NA
#> phyPLS-5 NA NA
#> phyPLS-6 NA NA
#> phyPLS-7 NA NA
#> phyPLS-8 NA NA
#> phyPLS-9 NA NA
#> phyPLS-10 NA NA
plot(ppls$x2, ppls$x) #ordination
#perform phylogenetic PLS between shape and size (for species means)
ppls <- pls_shapes(shapes = sp_shapes, X = sp_sizes, tree = tree)
#inspect results
names(ppls) #the contents of the resulting object
#> [1] "sdev" "rotation" "x" "x2" "center" "totvar"
exp_var(ppls) #variance explained by each axis
#> variance cummulative
#> phyPLS-1 75.10454 75.10454
#> phyPLS-2 NA NA
#> phyPLS-3 NA NA
#> phyPLS-4 NA NA
#> phyPLS-5 NA NA
#> phyPLS-6 NA NA
#> phyPLS-7 NA NA
#> phyPLS-8 NA NA
#> phyPLS-9 NA NA
#> phyPLS-10 NA NA
plot(ppls$x2, ppls$x) #ordination
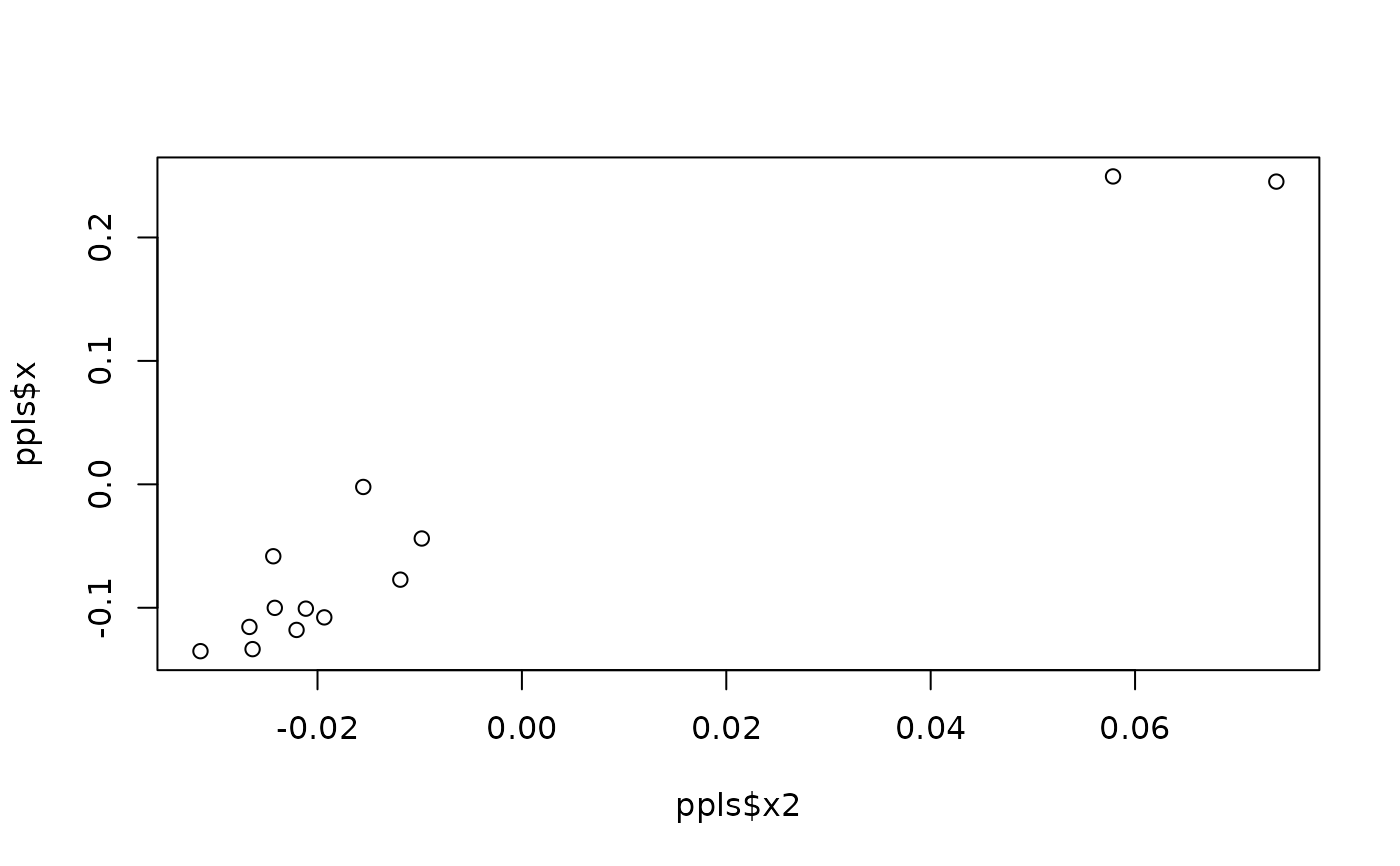 #perform phylogenetic PLS between shape and size with leave-one-out CV
ppls <- pls_shapes(shapes = sp_shapes, X = sp_sizes, tree = tree,
LOOCV = TRUE)
#inspect results
names(ppls) #the contents of the resulting object
#> [1] "sdev" "rotation" "x" "x2" "center" "totvar"
exp_var(ppls) #variance explained by each axis
#> variance cummulative
#> phyPLS-1 49.9805 49.9805
#> phyPLS-2 NA NA
#> phyPLS-3 NA NA
#> phyPLS-4 NA NA
#> phyPLS-5 NA NA
#> phyPLS-6 NA NA
#> phyPLS-7 NA NA
#> phyPLS-8 NA NA
#> phyPLS-9 NA NA
#> phyPLS-10 NA NA
plot(ppls$x2, ppls$x) #ordination
#perform phylogenetic PLS between shape and size with leave-one-out CV
ppls <- pls_shapes(shapes = sp_shapes, X = sp_sizes, tree = tree,
LOOCV = TRUE)
#inspect results
names(ppls) #the contents of the resulting object
#> [1] "sdev" "rotation" "x" "x2" "center" "totvar"
exp_var(ppls) #variance explained by each axis
#> variance cummulative
#> phyPLS-1 49.9805 49.9805
#> phyPLS-2 NA NA
#> phyPLS-3 NA NA
#> phyPLS-4 NA NA
#> phyPLS-5 NA NA
#> phyPLS-6 NA NA
#> phyPLS-7 NA NA
#> phyPLS-8 NA NA
#> phyPLS-9 NA NA
#> phyPLS-10 NA NA
plot(ppls$x2, ppls$x) #ordination