Back-calculate variables in their original scale from existing ordination data. Used internally.
rev_eigen(scores, vectors, center)Arguments
- scores
A matrix of scores resulting from an ordination.
- vectors
A matrix of eigenvector coefficients used to build the ordination.
- center
A vector with the mean values of the original variables used in the ordination.
Value
A 2-margins matrix of variables on their original scale.
See also
Examples
#load data and packages
library(Morpho)
library(geomorph)
data("tails")
#perform principal component analysis on tails data
pca <- prcomp(two.d.array(tails$shapes))
#transform the scores back to shapes
backshapes_mat <- rev_eigen(scores = pca$x,
vectors = pca$rotation,
center = pca$center)
backshapes_arr <- arrayspecs(backshapes_mat, k = 2, p = 9)
#compare
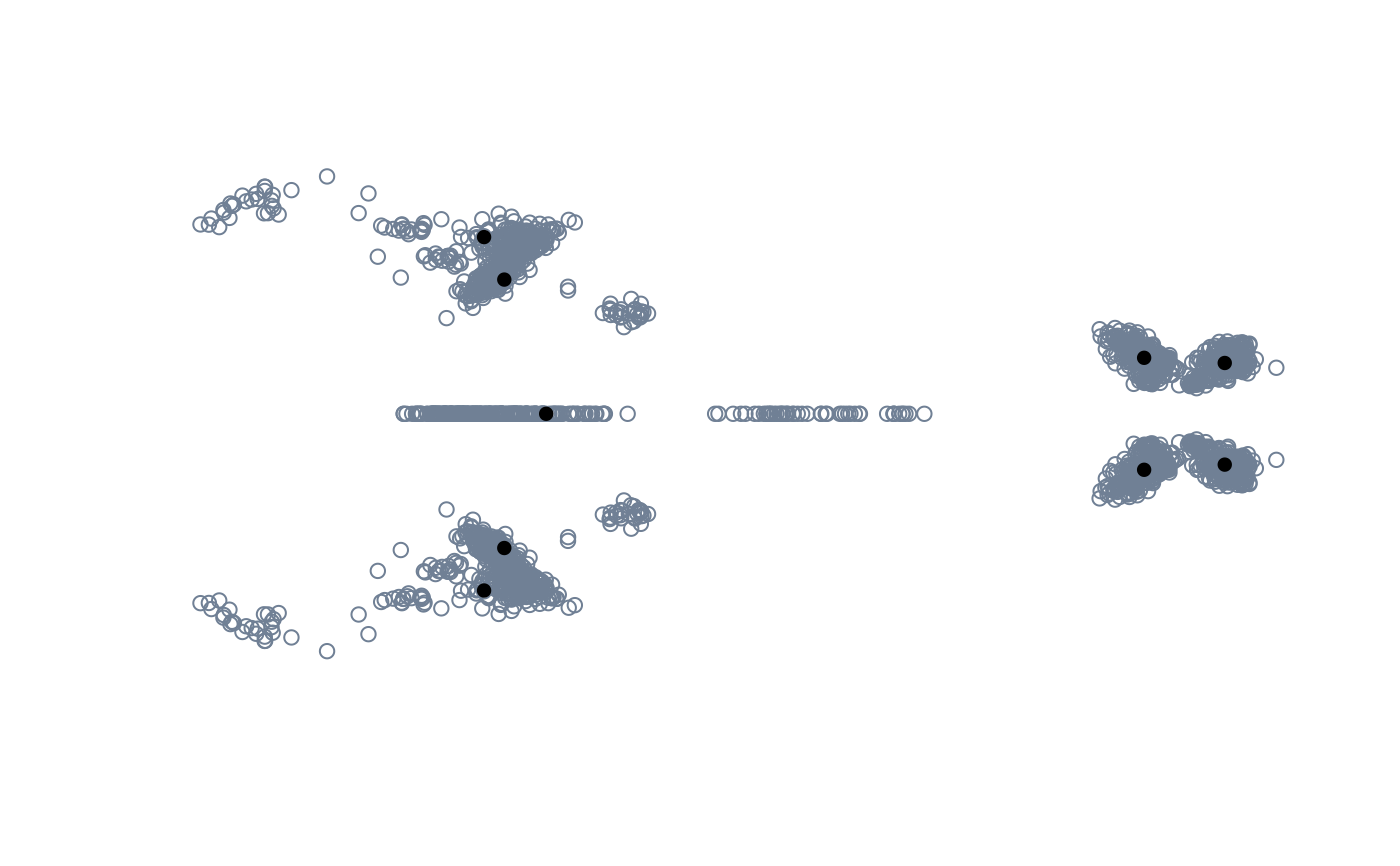
pile_shapes(tails$shapes)
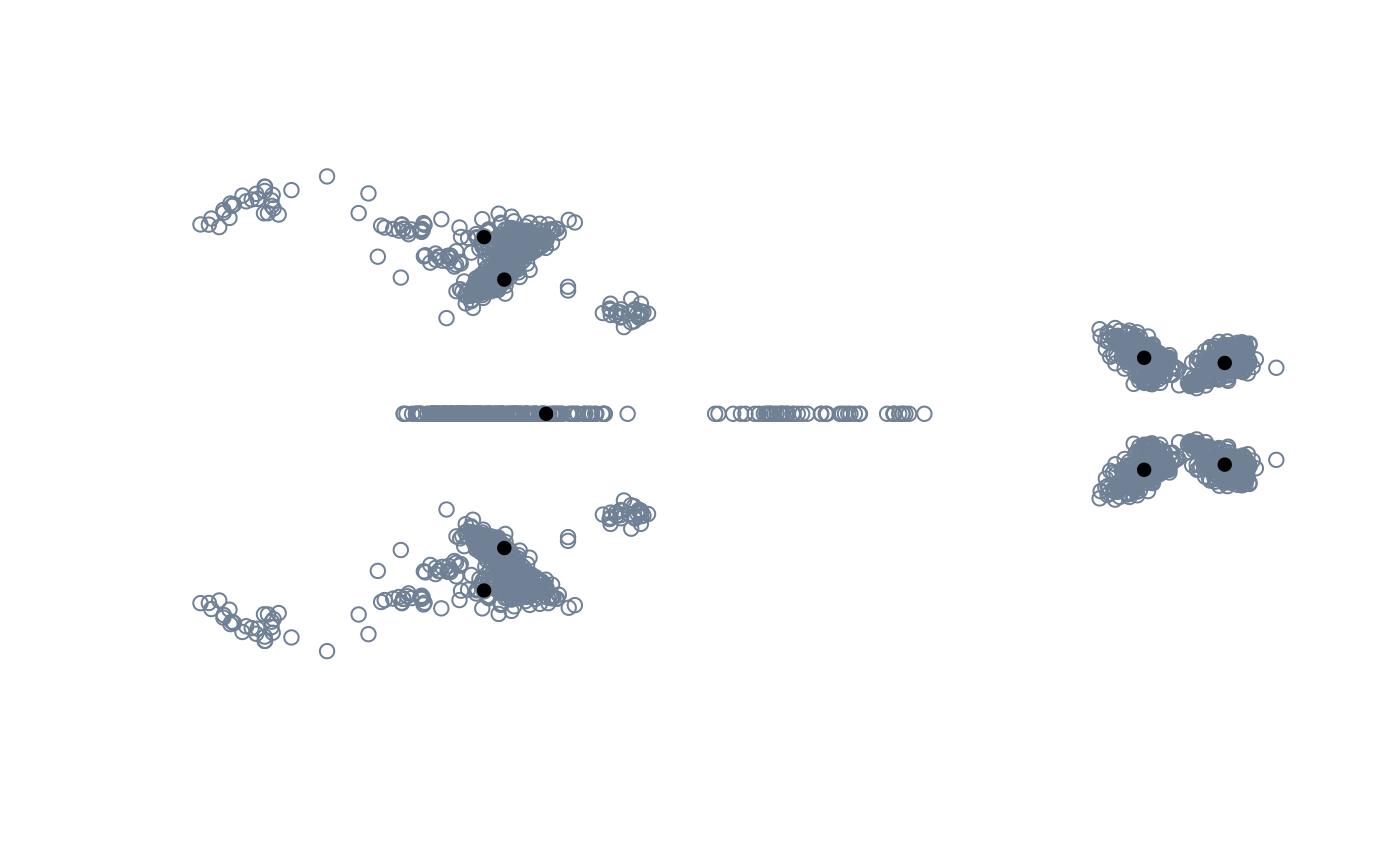 pile_shapes(backshapes_arr)
pile_shapes(backshapes_arr)
 #obtain shapes at the extremes of PC1
extshapes_mat <- rev_eigen(scores = range(pca$x[,1]),
vectors = pca$rotation[,1],
center = pca$center)
extshapes_arr <- arrayspecs(extshapes_mat, k = 2, p = 9)
#plot and compare
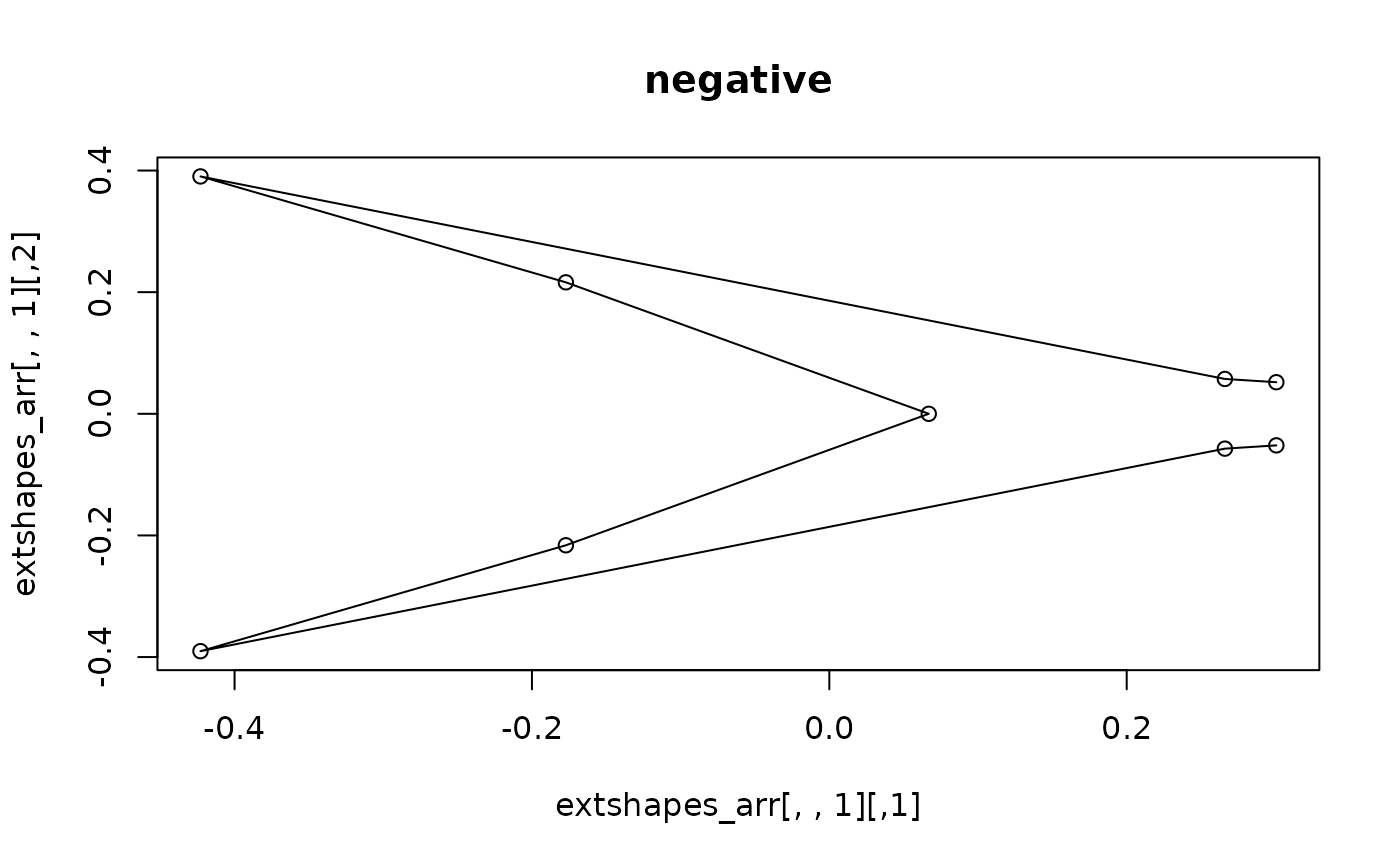
plot(extshapes_arr[,,1])
lineplot(extshapes_arr[,,1], tails$links) ; title("negative")
#obtain shapes at the extremes of PC1
extshapes_mat <- rev_eigen(scores = range(pca$x[,1]),
vectors = pca$rotation[,1],
center = pca$center)
extshapes_arr <- arrayspecs(extshapes_mat, k = 2, p = 9)
#plot and compare
plot(extshapes_arr[,,1])
lineplot(extshapes_arr[,,1], tails$links) ; title("negative")
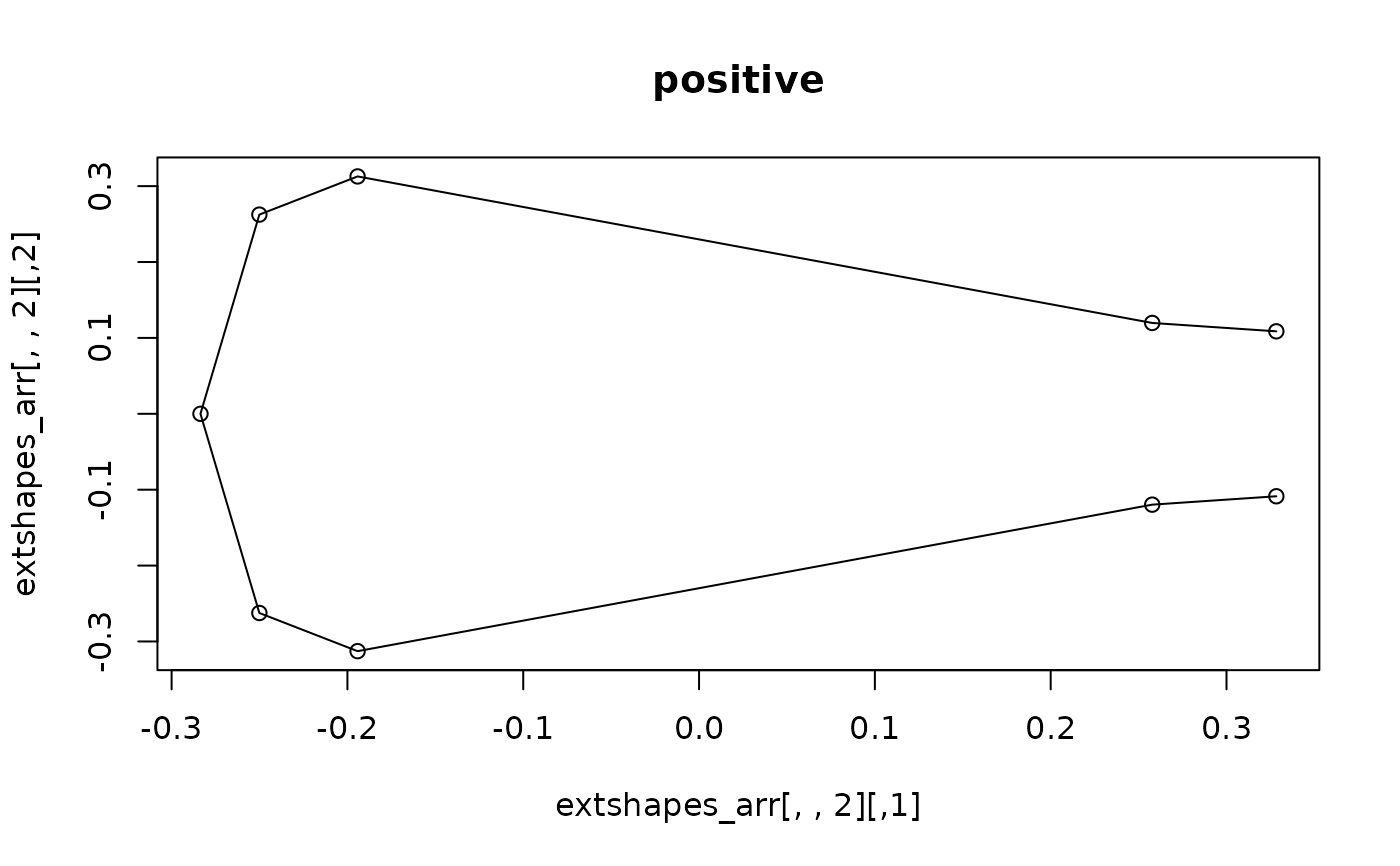
 plot(extshapes_arr[,,2])
lineplot(extshapes_arr[,,2], tails$links) ; title("positive")
plot(extshapes_arr[,,2])
lineplot(extshapes_arr[,,2], tails$links) ; title("positive")